Lateral gene transfer (LGT) is an important mechanism for genome diversification in microbial communities, including the human microbiome. While methods exist to identify LGTs from sequenced isolate genomes, identifying LGTs from community metagenomes remains an open problem. To address this, we developed WAAFLE: a Workflow to Annotate Assemblies and Find LGT Events.
User Manual || User Tutorial || Forum
Citation:
Profiling lateral gene transfer events in the human microbiome using WAAFLE
DOI: 10.1038/s41564-024-01881-w
1 Harvard T.H. Chan School of Public Health, Boston, MA, USA.
2 Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
3 Faculty of Computer Science, Dalhousie University, Halifax, Nova Scotia, Canada.
4 The Broad Institute of MIT and Harvard, Cambridge, MA, USA.
5 Department of Pharmacology, Dalhousie University, Halifax, Nova Scotia, Canada.
6 Harvard T.H. Chan School of Public Health, Boston, MA, USA. lnguyen24@mgh.harvard.edu.
7 Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA. lnguyen24@mgh.harvard.edu.
8 Harvard T.H. Chan School of Public Health, Boston, MA, USA. franzosa@hsph.harvard.edu.
9 The Broad Institute of MIT and Harvard, Cambridge, MA, USA. franzosa@hsph.harvard.edu.
#Contributed equally.
-
- Install the WAAFLE software:
$ conda install waafle -c biobakery- See here for detailed instructions on installing bioBakery conda recipes (e.g. setting channel priorities)
- See the WAAFLE manual for other installation options
- Download the WAAFLE BLAST database and taxonomy file:
- chocophlan2.tar.gz (4.3 GB)
- chocophlan2_taxonomy.tsv (<1 MB)
- Unpack the BLAST database:
$ tar xzfv chocophlan2.tar.gz
- Install the WAAFLE software:
conda, as suggested in the quick installation instructions above.- Python 3+ or 2.7+
- Python
numpy(tested with v1.13.3) - NCBI BLAST+ (tested with v2.6.0)
- bowtie2 (for performing read-level QC; tested with v2.2.3)
- You will need a multifasta file containing metagenomic contigs:
- Referred to as
contigs.fnabelow - Or download and try demo_contigs.fna
- Referred to as
- Search your contigs against the WAAFLE database:
$ waafle_search contigs.fna chocophlan2/chocophlan2- This creates
contigs.blastout(BLAST hits)
- Identify ORFs from your contigs and BLAST results:
$ waafle_genecaller contigs.blastout- This creates
contigs.gff(gene calls)
- Taxonomically classify contigs and find LGT events:
$ waafle_orgscorer contigs.fna contigs.blastout contigs.gff chocophlan2_taxonomy.tsv- This creates
contigs.no_lgt.tsv(single-clade contigs) - This creates
contigs.lgt.tsv(putative LGT events)
WAAFLE integrates gene sequence homology and taxonomic provenance to identify metagenomic contigs explained by pairs of microbial clades but not by single clades (i.e. putative LGTs). More specifically, for each locus in a contig, WAAFLE identifies the best hit to each species in a pangenome database. WAAFLE then looks for a species whose minimum per-locus score exceeds a lenient homology threshold (k1). If one or more species meet this criterion, then the contig is assigned to the species with the best average score. Otherwise, the process is repeated for pairs of species. If all per-locus scores for a pair of species exceed a stringent homology threshold (k2), then the contig is considered a putative LGT between those species.
Consider the following pair of examples:
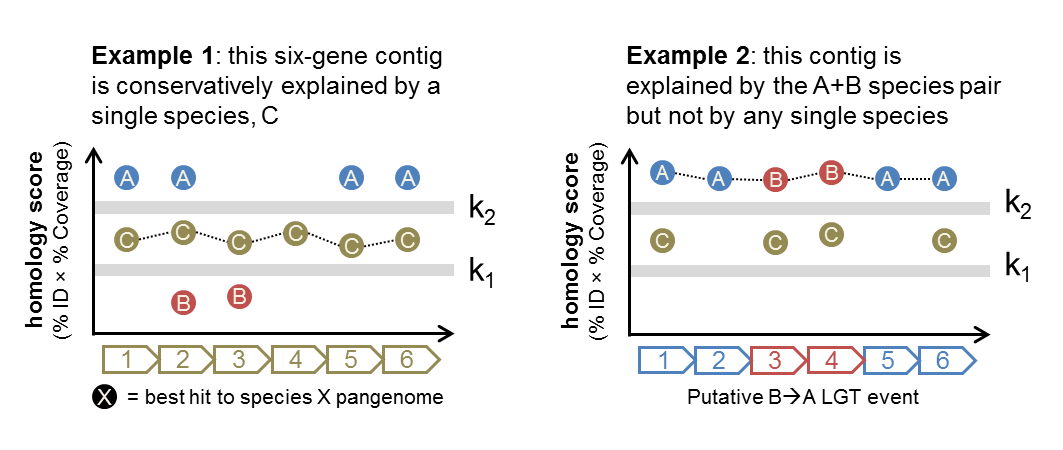
Both cases consider contigs with six protein-coding loci (determined from WAAFLE itself or an independent ORF-calling program such as Prodigal). In Example 1, genes from species C are able to explain all of the loci reasonably well (with scores exceeding k1). Hence, WAAFLE will report this contig as a one-species contig explained by species C.
In Example 2, no single species can explain all of the loci (the minimum score for each species is below k1). However, the pair of species A and B have strong hits (>k2) to all loci, and so WAAFLE concludes that this contig may represent an A+B LGT. Given the AABBAA synteny pattern, a B-to-A transfer would appear to be the more likely mechanism.
Note that in Example 2, if species C had hits to the 2nd and 5th loci that exceeded k1 (as in Example 1), WAAFLE’s algorithm would conservatively favor the weaker one-species explanation for the contig rather than invoking a two-species (LGT-based) explanation.
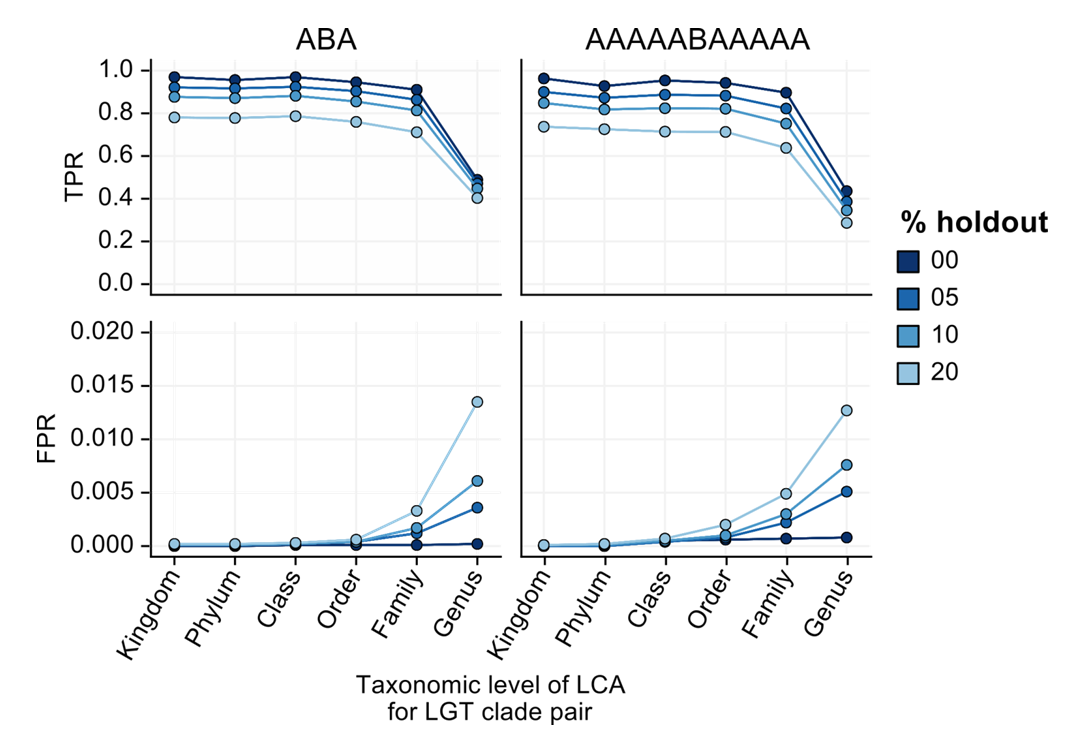
We evaluated WAAFLE on synthetic contigs with prespecified synteny patterns. Synthetic contigs were always assembled from individual genes drawn from a pair of genomes, A and B. When A and B represent two different species, the contig is considered a positive LGT for TPR calculation (the “level” of the LGT is given by the level of the LCA for A and B, with intra-genus LGTs being the lowest level). When A and B represent two strains of the same species, the contig is considered a negative for FPR calculation.
Even as fractions of the underlying species database were held out (from 0 to 20%), WAAFLE tended to remain >60% specific for LGTs at the family level or higher and >99% specific at all levels of taxonomic resolution.
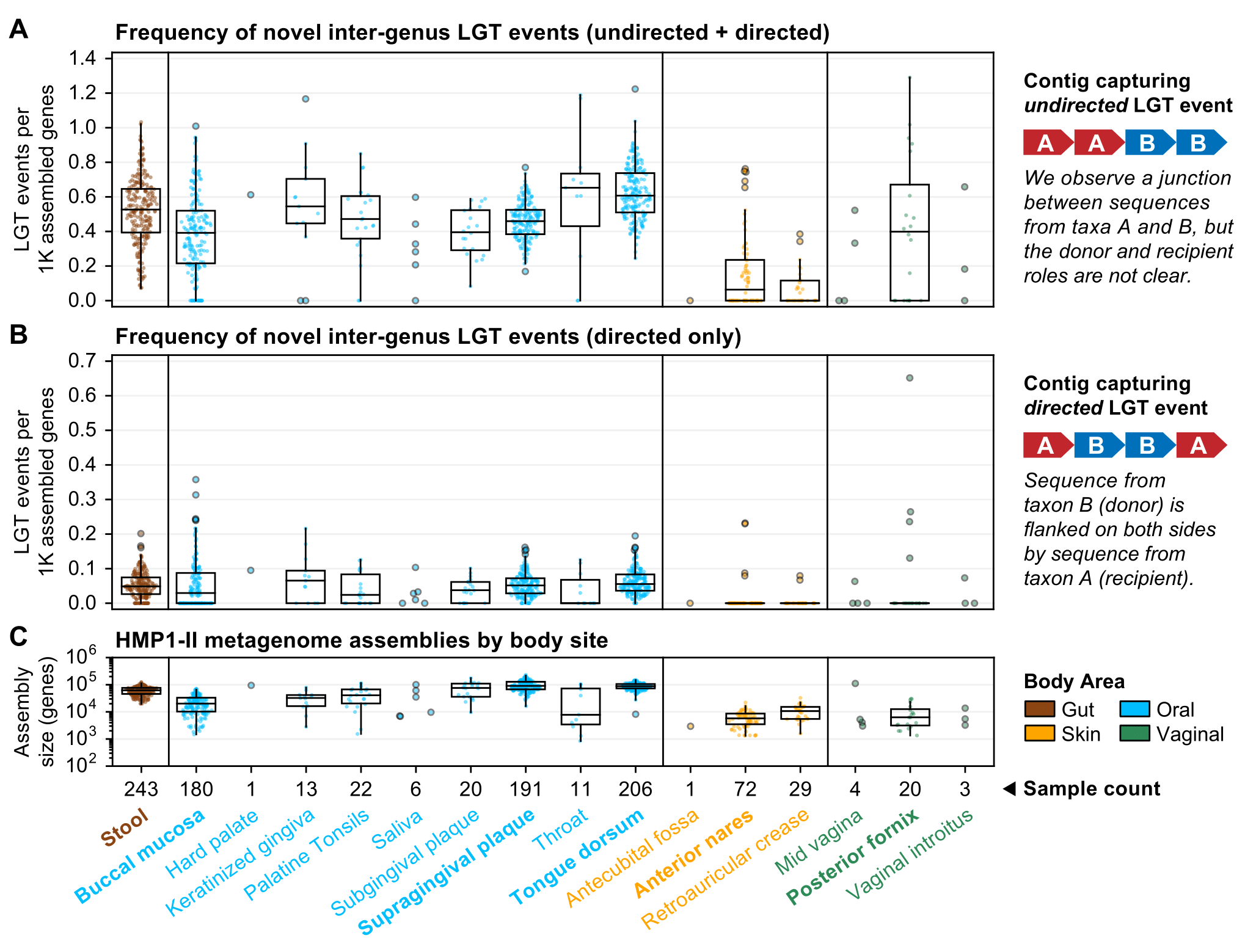
We are applying WAAFLE to quantify rates of LGT in the human microbiome using the HMP1-II dataset. Here, we report the LGT rates and assembly sizes at eight human body sites as sampled from at least 20 healthy adults. This analysis conservatively only counts LGTs with i) known directionality, ii) an LCA above the genus level, and iii) genus-level resolution or better.
WAAFLE databases (publication versions):
- WAAFLE nucleotide BLAST database of microbial pangenomes and corresponding taxonomy file adapted from the ChocoPhlAn 2.0 pangenome catalog underlying MetaPhlAn 2 and HUMAnN 2:
- chocophlan2.tar.gz (4.3 GB)
- chocophlan2_taxonomy.tsv (<1 MB)
Synthetic validation data
- Synthetic data (contigs and annotations) used in the evaluation of WAAFLE:
- synthetic_data.tar.gz (0.5 GB)
HMP1-II contigs and LGT profiles
- MEGAHIT assemblies of HMP1-II metagenomes (contig sets individually compressed within tar files):
- megahit.hmp1-ii.Anterior_nares.tar (0.4 GB)
- megahit.hmp1-ii.Buccal_mucosa.tar (2.6 GB)
- megahit.hmp1-ii.Posterior_fornix.tar (0.3 GB)
- megahit.hmp1-ii.Stool.tar (18 GB)
- megahit.hmp1-ii.Supragingival_plaque.tar (13 GB)
- megahit.hmp1-ii.Tongue_dorsum.tar (14 GB)
- megahit.hmp1-ii.Other.tar (2.0 GB)
- WAAFLE profiles of above HMP1-II assemblies, post-quality control:
- output.qcpass.hmp1-ii.tar.gz (3.4 GB)
HMP2 contigs and LGT profiles
-
- MEGAHIT assemblies of first-visit stool metagenomes from 26 HMP2 control subjects (contig sets individually compressed within tar file):
- megahit.hmp2.tar (0.5 GB)
- WAAFLE profiles of above HMP2 assemblies, post-quality control:
- output.qcpass.hmp2.tar.gz (<1 MB)
- MEGAHIT assemblies of first-visit stool metagenomes from 26 HMP2 control subjects (contig sets individually compressed within tar file):
