Genetic and genomic variation among microbial strains can dramatically influence their phenotypes and environmental impact, including on human health. However, inferential methods for quantifying these differences have been lacking. Strain-level metagenomic profiling data has several features that make traditional statistical methods challenging to use, including high dimensionality, extreme variation among samples, and complex phylogenetic relatedness. We present Anpan, a set of quantitative methods addressing three key challenges in microbiome strain epidemiology. First, adaptive filtering designed to interrogate microbial strain gene carriage is combined with linear models to identify strain-specific genetic elements associated with host health outcomes and other phenotypes. Second, phylogenetic generalized linear mixed models are used to characterize the association of sub-species lineages with such phenotypes. Finally, random effects models are used to identify pathways more likely to be retained or lost by outcome-associated strains. We validated our methods by simulation, showing that we achieve more accurate effect size estimation and a lower false positive rate compared to alternative methodologies. We then applied our methods to a dataset of 1,262 colorectal cancer patients, identifying functionally adaptive genes and strong phylogenetic effects associated with CRC status, sometimes complementing and sometimes extending known species-level microbiome CRC biomarkers. Anpan’s methods have been implemented as a publicly available R library to support microbial community strain and genetic epidemiology in a variety of contexts, environments, and phenotypes.
User manual || Forum || Tutorial
Citation:
Andrew R. Ghazi1,2, Kelsey N. Thompson1,2,4, Amrisha Bhosle1,2,4, Zhendong Mei3, Yan Yan1, Fenglei Wang5, Kai Wang6, Eric A. Franzosa1,2,4, Curtis Huttenhower1,2,4,7
Quantifying Metagenomic Strain Associations from Microbiomes with Anpan (In-prep)
1 Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA
2 Infectious Disease and Microbiome Program, Broad Institute of MIT and Harvard, Cambridge, MA, USA
3 Channing Division of Network Medicine, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA
4 The Harvard Chan Microbiome in Public Health Center, Boston, MA, USA
5 Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA
6 Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA
7 Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, MA, USA
Once you have the dependencies, you can install anpan from github with:
remotes::install_github("biobakery/anpan")If you would like to read the walkthrough vignette, you will need to set the build_vignettes = TRUE argument in theinstall_github() command, at which point you can view the vignette with vignette("anpan_tutorial", package = 'anpan'). There is also a static (and probably not perfectly up-to-date) version of the tutorial available on the biobakery wiki GitHub page.
anpan depends on R >= 4.1 and the following R packages, most of which are available through CRAN (the exception being cmdstanr):
install.packages(c("ape",
"data.table",
"dplyr",
"fastglm",
"furrr",
"ggdendro",
"ggnewscale",
"ggplot2",
"loo",
"patchwork",
"phylogram",
"posterior",
"progressr",
"purrr",
"R.utils",
"remotes",
"stringr",
"tibble",
"tidyselect")) # add Ncpus = 4 to go faster
install.packages("cmdstanr", repos = c("https://mc-stan.org/r-packages/", getOption("repos")))If the cmdstanr installation doesn’t work you can find more detailed instructions at this link. Once you’ve installed cmdstanr, you will need to use it to install CmdStan itself:
library(cmdstanr)
check_cmdstan_toolchain()
install_cmdstan(cores = 2)
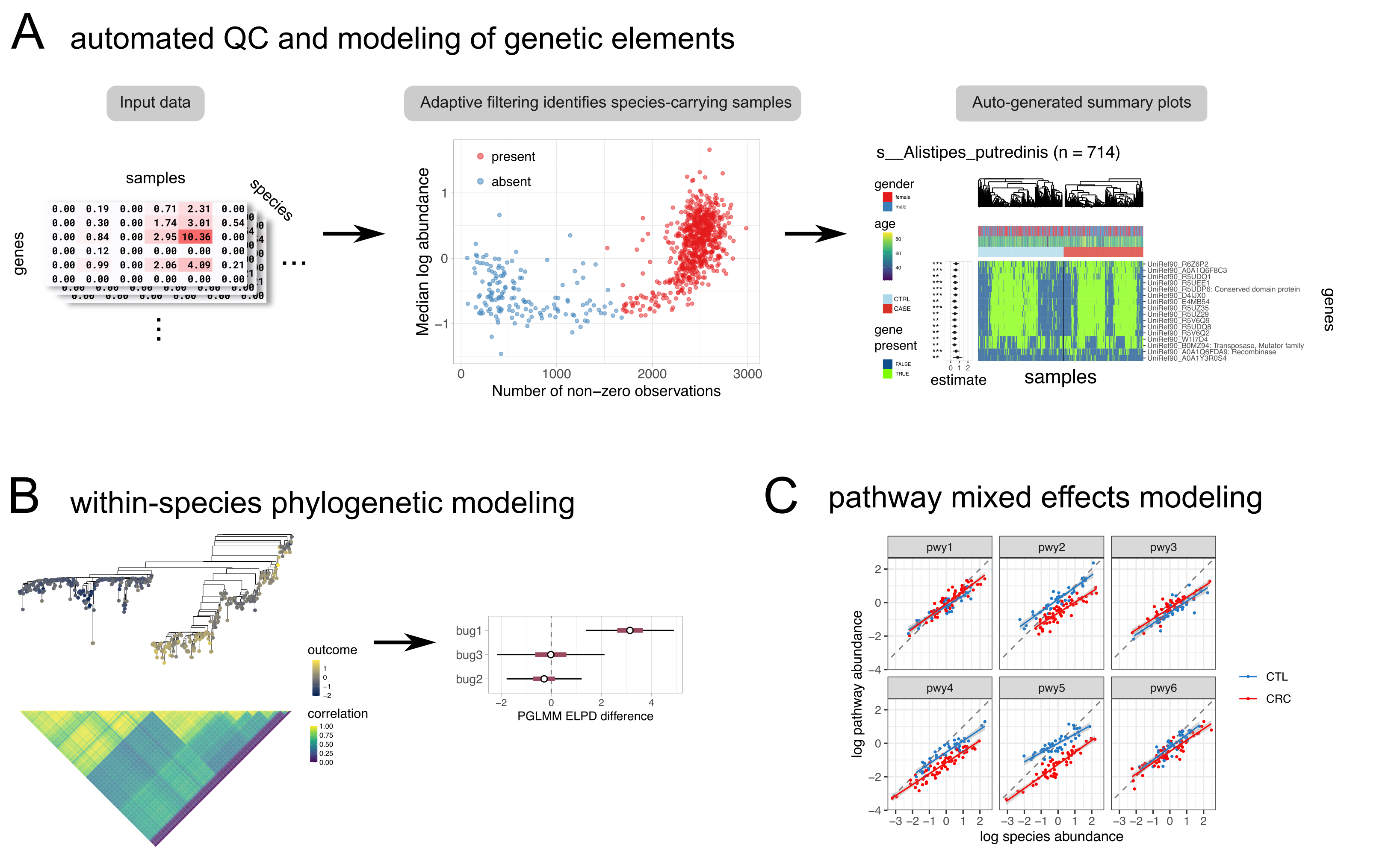
You can filter large uniref tables and look for associations with outcomes (while controlling for covariates) withanpan_batch.
library(anpan)
anpan_batch(bug_dir = "/path/to/gene_families/",
meta_file = "/path/to/metadata.tsv",
out_dir = "/path/to/output",
annotation_file = "/path/to/annotation.tsv", #optional, used for plots
filtering_method = "kmeans",
model_type = "fastglm",
covariates = c("age", "gender"),
outcome = "crc",
plot_ext = "pdf",
save_filter_stats = TRUE)This will run an anpan batch analysis for each functional profile present in bug_dir (there should be one input file per bug). For each bug, anpan:
- reads in the metadata (containing the specified outcome and covariates) and the functional profile
- applies adaptive filtering to discard samples where the species was likely not present (based on low abundance and few non-zero observations) based on the sample’s functional profile.
- fits a model to the filtered data:
model_type = "fastglm"ormodel_type = "glm"will fit regressions to each gene in the input file separately (using the specified outcome distribution), then apply FDR correction to the p-values associated with each gene’s presence. The regression for each individual gene follows the form:outcome ~ covariate1 + covariate2 + ... + gene_presencemodel_type = "horseshoe"fits a single regression model with the specified outcome distribution, weak priors on the specified covariates, and a regularized horseshoe prior on the presence/absence of all genes associated with the bug. The regression follows the form:outcome ~ covariate1 + covariate2 + ... + gene1_presence + gene2_presence + gene3_presence + ...including all of the specified covariates and (typically thousands of) genes detected in the bug.
- then writes results to the output directory, including tables of regression coefficients, per-bug plots of the top hits, and filter statistics.
You can run a PGLMM with the anpan_pglmm function. This fits a model where the covariance matrix of outcomes is determined from the phylogenetic tree. Right now the only usable outcome distributions are family = "gaussian" and family = "binomial". If family = "gaussian", the reg_noise parameter can be used to regularize the ratio of the noise parameters with the following prior distribution:
meta_file = "/path/to/metadata.tsv" tree_file = "/path/to/file.tre" anpan_pglmm(meta_file, tree_file, trim_pattern = "_bowtie2", covariates = NULL, plot_cov_mat = TRUE, outcome = "age", verbose = TRUE, cores = 4)
There is a corresponding anpan_pglmm_batch() function that can be pointed to a directory of .tree files with the tree_dir parameter.
