Kneaddata
KneadData is a tool designed to perform quality control on metagenomic sequencing data, especially data from microbiome experiments. In these experiments, samples are typically taken from a host in hopes of learning something about the microbial community on the host. However, metagenomic sequencing data from such experiments will often contain a high ratio of host to bacterial reads. This tool aims to perform principled in silico separation of bacterial reads from these “contaminant” reads, be they from the host, from bacterial 16S sequences, or other user-defined sources.
User Manual || User Tutorial || Forum
Requirements
- Trimmomatic (version == 0.33) (automatically installed)
- Bowtie2 (version >= 2.2) (automatically installed)
- Python (version >= 2.7)
- Java Runtime Environment
- TRF (optional)
- Fastqc (optional)
- SAMTools (only required if input file is in BAM format)
- Memory (>= 4 Gb if using Bowtie2, >= 8 Gb if using BMTagger)
- Operating system (Linux or Mac)
Optionally, BMTagger can be used instead of Bowtie2.
The executables for the required software packages should be installed in your $PATH. Alternatively, you can provide the location of the Bowtie2 install ($BOWTIE2_DIR) with the following KneadData option “–bowtie2 $BOWTIE2_DIR”.
Getting started
Installation
Before installing KneadData, please install the Java Runtime Environment (JRE). First download the JRE for your platform. Then follow the instructions for your platform: Linux 64-bit or Mac OS. At the end of the installation, add the location of the java executable to your $PATH.
- Install the KneadData software
$ pip install kneaddata- This command will automatically install Trimmomatic and Bowtie2. To bypass the install of dependencies, add the option “”–install-option=’–bypass-dependencies-install'”.
- If you do not have write permissions to ‘/usr/lib/’, then add the option “–user” to the install command. This will install the python package into subdirectories of ‘$HOME/.local/’. Please note when using the “–user” install option on some platforms, you might need to add ‘$HOME/.local/bin/’ to your $PATH as it might not be included by default. You will know if it needs to be added if you see the following message
kneaddata: command not foundwhen trying to run KneadData after installing with the “–user” option.
- Download the human reference database (approx. size = 3.8 GB)
$ kneaddata_database --download human_genome bowtie2 $DIR- When running this command, $DIR should be replaced with the full path to the directory you have selected to store the database.
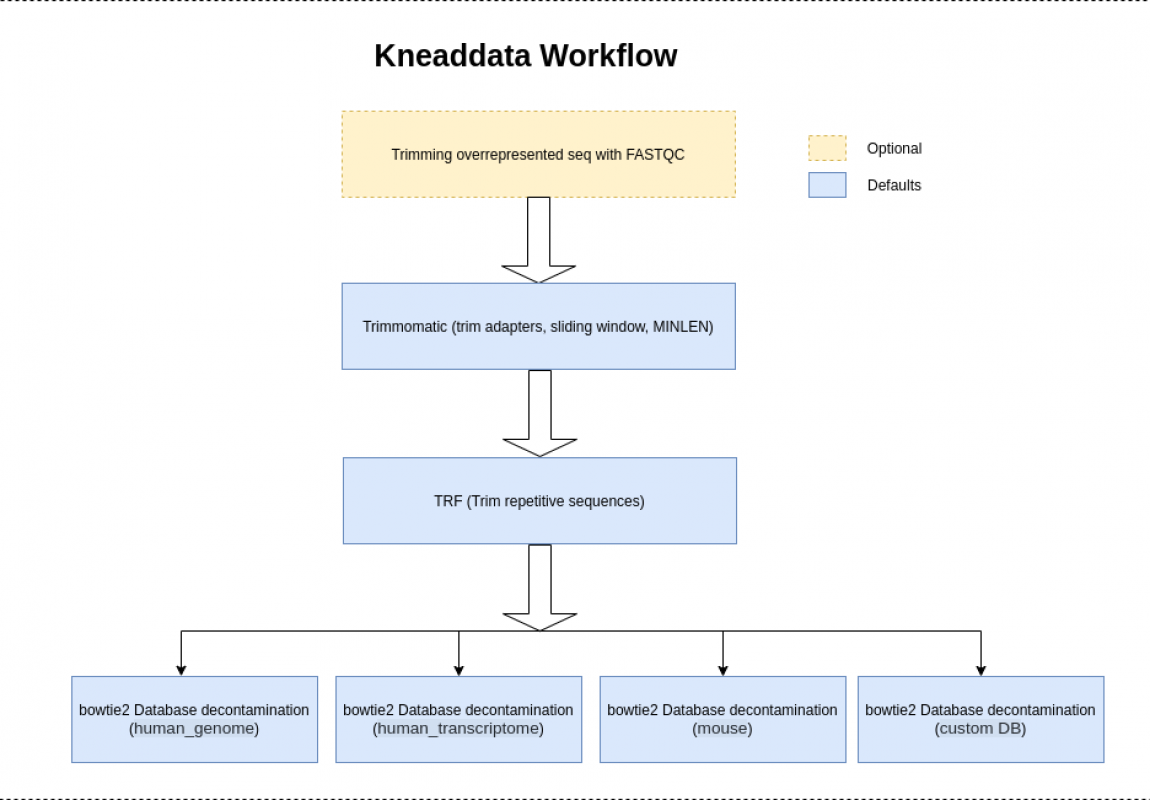
How to Run
Basic usage
Kneaddata >=v0.11.0
Single-End Inputs:
$ kneaddata --unpaired $UNPAIRED --reference-db $DATABASE --output $OUTPUT_DIR
Paired-End Inputs:$ kneaddata --input1 $INPUT1 --input2 $INPUT2 --reference-db $DATABASE --output $OUTPUT_DIR
$UNPAIRED = a single end fastq file (can be gzipped) or a SAM/BAM formatted file
$INPUT1 = R1 pair-end fastq file (can be gzipped) or a SAM/BAM formatted file
$INPUT2 = R2 pair-end fastq file (can be gzipped) or a SAM/BAM formatted file
$DATABASE = the index of the KneadData database $OUTPUT_DIR = the output directory
For paired end reads, add a second input argument “–input $INPUT2” (with $INPUT2 replaced with the second input file). Also please note that more than one reference database can be provided in the same manner by using multiple database options (for example, “–reference-db $DATABASE1 –reference-db $DATABASE2”). Providing a database is optional. If a database is not provided the step of testing for contaminant sequences from a reference database will be bypassed.
Four types of output files will be created (where $INPUTNAME is the basename of $INPUT):
The final file of filtered sequences after trimming
$OUTPUT_DIR/$INPUTNAME_kneaddata.fastq
The contaminant sequences from testing against a database (with this database name replacing $DATABASE and “bowtie2” or “bmtagger” replacing $SOFTWARE)
$OUTPUT_DIR/$INPUTNAME_kneaddata_$DATABASE_$SOFTWARE_contam.fastq
The log file from the run
$OUTPUT_DIR/$INPUTNAME_kneaddata.log
The fastq file of trimmed sequences
$OUTPUT_DIR/$INPUTNAME_kneaddata.trimmed.fastq- Trimmomatic is run with the following arguments by default “SLIDINGWINDOW:4:20 MINLEN:70”. The minimum length is computed as 70 percent of the length of the input reads. To change the Trimmomatic arguments, use the option “–trimmomatic-options”.
If there is more than one reference database, then more than one file of contaminant sequences will be written. If running with two input files, each type of fastq output file will be created for each one of the pairs of the input files. If running with the TRF step, an additional set of files with repeats removed will be written.
Demo run
The examples folder in the KneadData source archive contains a demo input file and a demo database. The input file is in fastq format.
(>=v0.11.0)$ kneaddata --unpaired examples/demo.fastq --reference-db examples/demo_db --output kneaddata_demo_output
(<=v0.10.0)$ kneaddata --input examples/demo.fastq --reference-db examples/demo_db --output kneaddata_demo_output
This will create four output files:
kneaddata_demo_output/demo_kneaddata.fastqkneaddata_demo_output/demo_kneaddata_demo_db_bowtie2_contam.fastqkneaddata_demo_output/demo_kneaddata.logkneaddata_demo_output/demo_kneaddata.trimmed.fastq
