MaAsLin3 is an R package for efficiently determining multivariable associations between clinical metadata and microbial meta-omics features. Relative to MaAsLin 2, MaAsLin 3 introduces the ability to quantify and test for both abundance and prevalence associations while accounting for compositionality. By incorporating generalized linear models, MaAsLin 3 accomodates most modern epidemiological study designs including cross-sectional and longitudinal studies.
For more information on the technical aspects:
User manual || Tutorial || Forum
Citation:
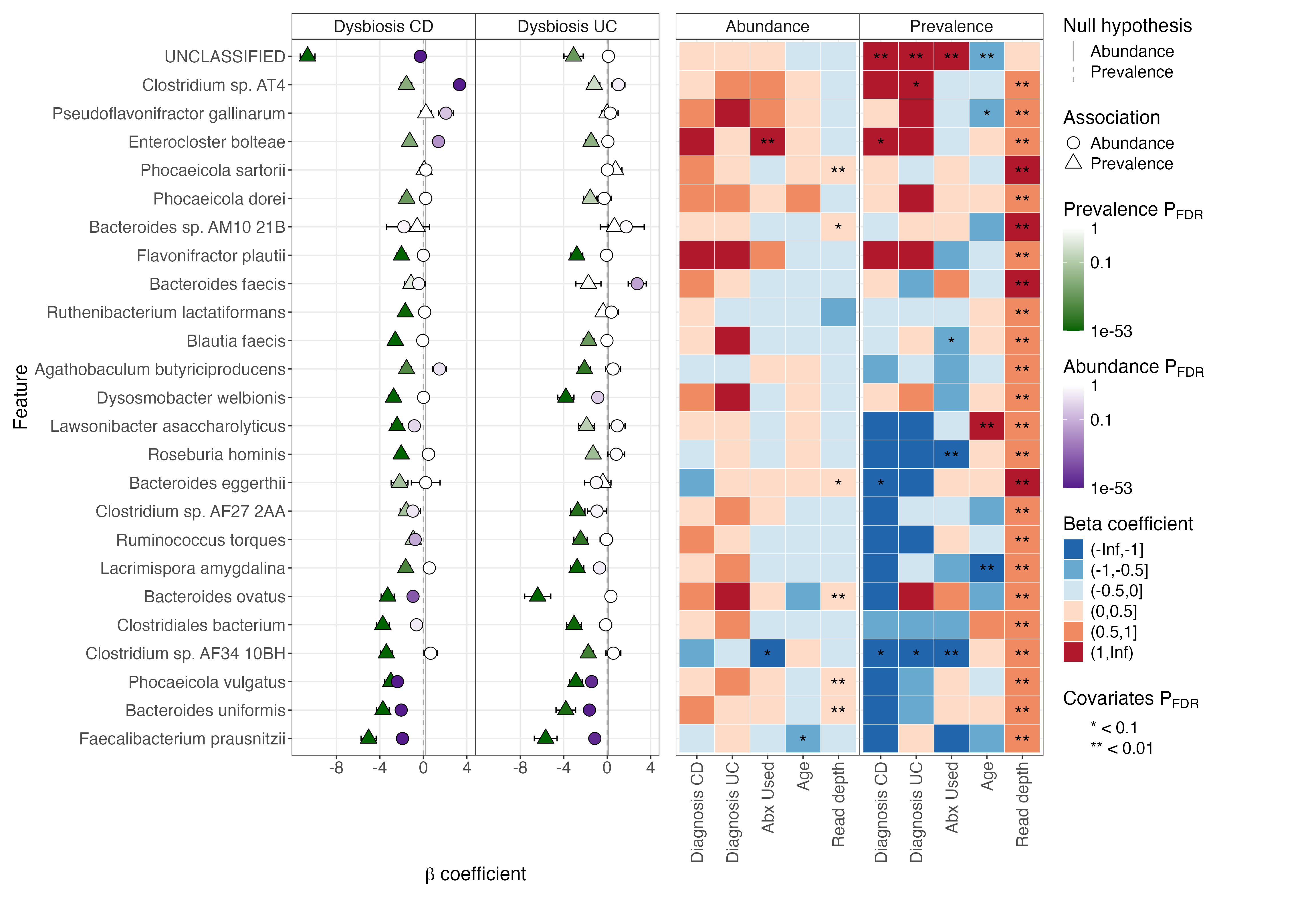
- Modeling both abundance and prevalence (presence/absence) associations
- Accounting for compositionality with spike-ins, total abundance scaling, or a median comparison technique
- Expanding modeling options with generalized formula parsing
- Extending inference with contrast tests for ordered predictors and ANOVA-style tests for differences among groups
- R software
- R CRAN packages: dplyr, pbapply, lmerTest, parallel, lme4, optparse, logging, data.table, multcomp, ggplot2, RColorBrewer, patchwork, scales, rlang, hash, matrixStats, tibble, ggnewscale, kableExtra
MaAsLin3 is an R package that can be run as a function within R or from the command line. It is available through GitHub with the package devtoools.
To install MaAsLin 3, run the following commands in R:
library("devtools")
install_github("biobakery/maaslin3")
library(maaslin3)
